In six subprojects (SP) we are investigating different questions on multitrophic interactions with the overall objective to identify mechanisms underlying the relationships of biodiversity with ecosystem functions across trophic levels.
Z1: Administrative and scientific coordination
Spokesperson:
Alexandra-M. Klein, University of Freiburg
Project Coordinator:
Finn Rehling, University of Freiburg
Chinese Coordinator:
Chao-Dong Zhu, Institute of Zoology, Chinese Academy of Sciences, Beijing

SP1: Wood decomposition and decomposer interactions
Objectives
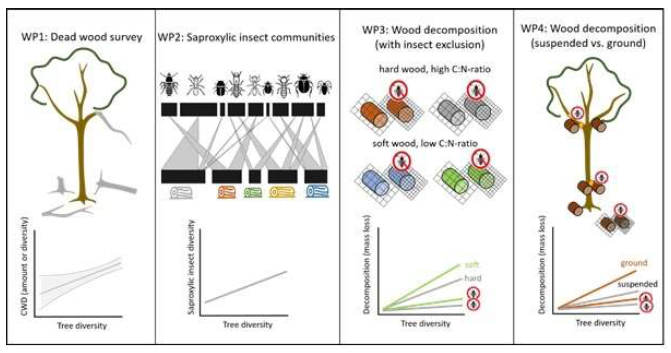
The overall objectives of SP1 are to characterize the bottom up effects of tree diversity on (1) the amount and diversity of naturally occurring dead wood to assess resource availability and quality for saproxylic insects and nutrient input into soil along this decomposition pathway, (2) the (functional trait) diversity of saproxylic insect communities in naturally occurring dead wood and how these are shaped indirectly by tree diversity through changes in biotic interactions that may influence community assembly, (3) functional traits of saproxylic insects in dead wood and their effect on wood decomposition rate using experimentally exposed dead wood with and without insect exclusion, (4) comparison of dead wood colonization and decomposition rate of suspended dead wood with dead wood on the ground. Our work will be organized accordingly into four work packages (WP) following the main aims (see Fig. 1 below).
Researchers
Principle Investigators:
Heike Feldhaar, University of Bayreuth
Simon Thorn, University of Würzburg
Doctoral Researcher:
Matteo Dadda, University of Bayreuth
Chinese Partner:
Arong Luo, Institute of Zoology, Chinese Academy of Sciences, Beijing
SP2: Soil-plant interactions and stoichiometry
Objectives
This subproject aims to test interrelations between trophic interactions, tree diversity,
and soil erosion with multi-element stoichiometry of resources and different trophic levels
(plants as primary producers and soil microorganisms as consumers). Additionally, the project
will conduct a full soil data baseline (all major plant nutrients and soil properties) as a courtesy
to all projects involved.
Our hypotheses are:
- Soil erosion redistributes nutrients along slopes and differentiates the nutrient pattern in soils available to soil microorganisms and trees. Since C and N are transported in the liquid phase and P bond to soil particles, the stoichiometry of the eroded material changes during transport and can translate into higher trophic levels via soil microorganisms and tree roots.
- Eroded parts show lower nutrient availability and foster nutrient recycling while deposition areas are dominated by nutrient overyielding and nutrient uptake. In contrast, diverse tree mixtures mitigate soil erosion and offset topographic differences along slopes and balance the trade-off between nutrient uptake and nutrient recycling.
- Soil microorganisms both recycle and take up nutrients more efficiently (= ‘nutrient overyielding’) in soil of diverse tree mixtures. However, nutrient overyielding is the dominant underlying mechanism and controls microbial stoichiometry.
- Similarly, trees both recycle and take up nutrients more efficiently in diverse tree mixtures. In contrast to soil microorganisms, nutrient recycling dominates as the underlying mechanism and thus, drives tree litter stoichiometry. Consequently, tree diversity effects on the stoichiometry of different trophic levels are decoupled.
Researchers
Principle Investigators:
Yvonne Oelmann, University of Tübingen
Thomas Scholten, University of Tübingen
Steffen Seitz, University of Tübingen
Doctoral Researcher:
Haikuo Zhang, University of Tübingen
Chinese Partners:
Yu Lian, Institute of Botany, Chinese Academy of Sciences, Peking
Naili Zhang, Beijing Forestry University
SP3: Plant-arthropod food webs, stoichiometry and functions
Objectives
By collecting data on community composition, consumption rates, body mass and elemental contents of arthropods as well as direct feeding links at higher trophic levels, we aim at predicting the responses of the multitrophic plant-herbivore-predator system to tree diversity loss. We specifically extend previous work by including three trophic levels (trees, herbivorous arthropods and predatory arthropods) and by linking multitrophic communities and their interactions with biomass distributions and stoichiometry to enable the analysis of trophic pyramids and energy-based networks. We will also record tritrophic species-level networks for selected tree species. We will use novel methods in the BEF-China Experiment (stoichiometry, gut content barcoding in collaboration with our Chinese collaborators) and will collect additional arthropod groups from other vegetation strata using multiple collection methods.
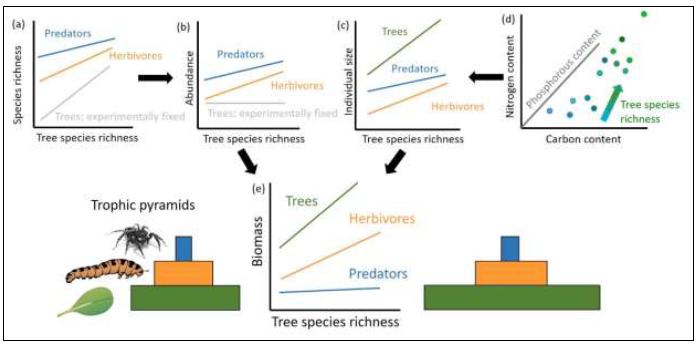
Based on the results of previous studies, we expect that tree diversity strongly affects consumer communities in various ways and hypothesise that (1) tree diversity effects on abundance and species richness decrease with increasing trophic level and this leads -together with changes in intra- and interspecific body size distributions of herbivores and predators – to shifts in trophic pyramids along the tree diversity gradient (Fig. 1). We further hypothesise that (2) intraspecific changes in carbon:nutrient ratios of herbivores and predators can be observed with increasing tree diversity (Fig. 1d), reflecting increased nutrient availability in plant tissues (within and across species) in more diverse plant communities. (3) Multitrophic plant-herbivore-predator energy networks are expected to show higher stocks (standing biomass) and fluxes (herbivory, predation) across trophic levels at higher tree diversity (Fig. 2a, d). In addition, we hypothesise that (4) structural properties of highly resolved plant-herbivore-predator networks provide more detailed mechanistic explanations of how changes in plant diversity affect ecosystem functioning.
Researchers
Principle Investigators:
Jana Petermann, University of Salzburg
Andreas Schuldt, University of Göttingen
Doctoral Researcher:
Mareike Mittag, University of Salzburg
Chinese Partners:
Douglas Chesters, Institute of Zoology, Chinese Academy of Sciences, Beijing
Ming-Qiang Wang, Chengdu Institute of Biology, Chinese Academy of Sciences, Chengdu
SP4: Cavity-nesting Hymenoptera and multi-trophic interactions
Objectives
Subproject 4 aims to distinguish tree diversity driven bottom up functions, acting on quantitative interaction networks of cavity-nesting Hymenoptera across trophic levels, including their food resources and natural enemies and tests drivers of cavity-nesting community establishment under natural conditions in dead wood as alternative nests. We will focus on separated overarching objectives in three work packages (WP):
Forest succession and the effects of tree diversity in multi-trophic interactions
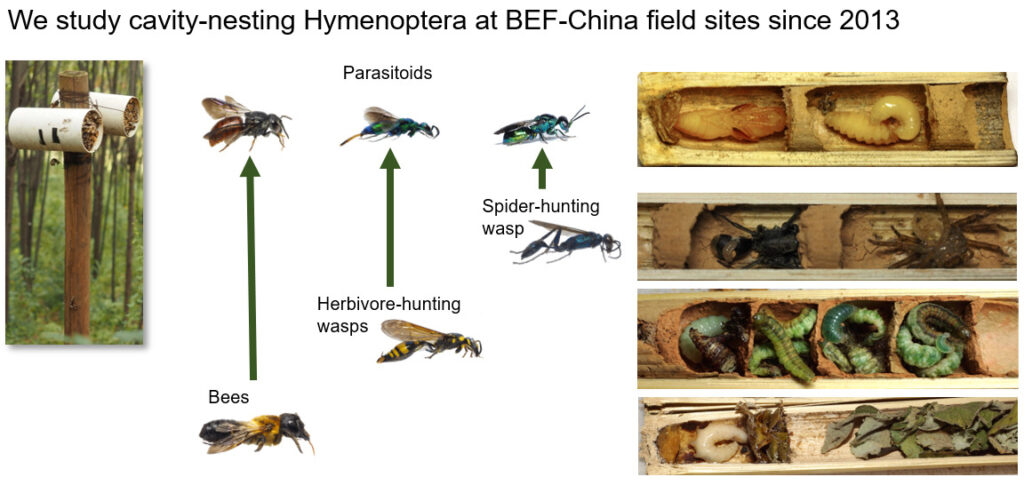
WP1 will address how tree diversity alters multi-trophic cavity-nesting Hymenoptera communities and their interactions with parasitoids. This first WP will study, for the first time, the influences of tree diversity along temporal changes of forest succession over a period of 10 years (2013-2024) at the 64 VIP plots. We will additionally analyse the relative importance of the different tree diversity components (taxonomic, functional and phylogenetic tree diversity) on cavity-nesting bees, wasps and their natural enemies in a controlled diversity gradient, installing, for the first time, reed nests at all 300 core plots. WP1 will test two hypotheses: (1) Tree diversity changes cavity-nesting communities, but only starting at the point after canopy closure with forest succession. This assumption is based on our comparison of cavity-nesting communities at the experimental sites during the first years of plot establishment with established old-growth forest plots. (2) Tree functional diversity explains higher complementarity of resources than taxonomic and phylogenetic tree diversity and should therefore be a strong predictor of the diversity of bee/wasp-parasitoid interactions. Traits affecting functions are not only related to bee and wasp feeding but also to nesting such as tree resin, which is beneficial for bee health.
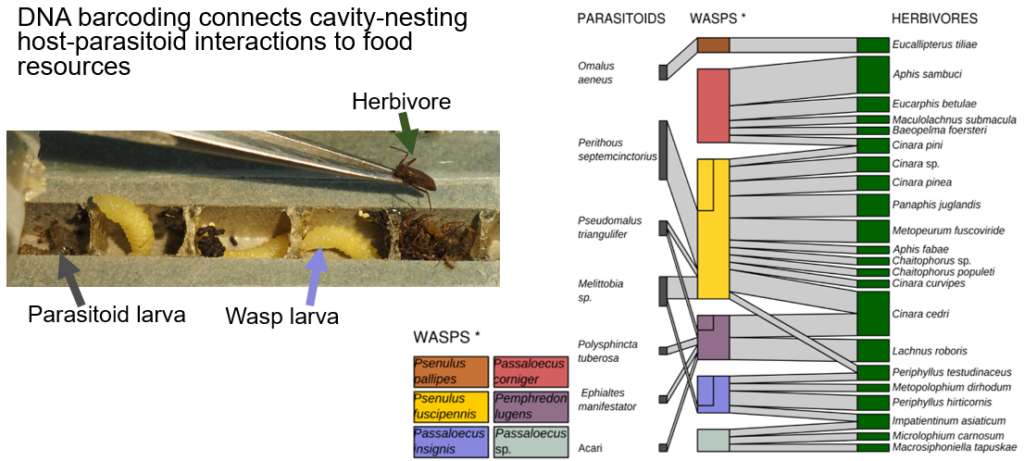
Multi-trophic network extension facilitated by DNA barcoding to link wasps to their herbivorous food prey
WP2 will extend the parasitoid-host interactions so far studied with reed nests to prey and food resources of hosts. This provides the unique opportunity to establish a mechanistic interaction link to the experimentally controlled trophic level of forest trees. Observed multi-trophic interactions of more than two trophic levels involving primary producers are lacking across forest BEF-studies worldwide (except for investigations on trophobiotic tree-ant-Hemiptera interactions at the BEF-China sites when trees were small and leaves accessible). With the identification of herbivorous prey, we aim to establish the feeding links between herbivore-hunting wasps with trees (together with SP3). Additionally, bee pollen will be directly attributed to flowering trees. We will establish the methodology of continuous prey identifications using barcoding and ultimately metabarcoding. As this is methodologically challenging, we will establish a DNA barcoding approach optimized for high-throughput sequencing (e.g. MinION and Illumina novaseq) to study samples taken only at the subset of VIP-plots. Given the challenging data analyses of Next Generation Sequencing (NGS) data, we will rely and refine already approved data analysis pipelines and develop a reference database comprising newly generated DNA libraries and barcodes as basis for the parallel establishment of the metabarcoding approach. WP2 will test two hypotheses: (1) High-throughput barcode analysis leads to high taxonomic resolution and is needed to establish a reliable metabarcoding approach for animal and plant tissue that leads to identifications at family, genus and species levels. (2) Tree diversity explains the degree of specialisation of bees and wasps but this pattern dampens with trophic-distance to the tree food resources.
Linking cavity-nesting bees, wasps and parasitoids to dead wood
WP3 will use a new cavity-nesting model (already tested in urban gardens in Germany) to link the trophic interactions of Hymenoptera to dead wood (coarse woody debris) and therefore to decomposers and ants. At each VIP plot of both study sites, we will install dead wood with cavities to simulate natural conditions for cavity-nesting Hymenoptera and dead wood decomposers. By comparing exclusion of ants to ant accessing dead wood, and comparing dead wood with drilled cavities to non-prepared dead wood (SP1), we study the competition and synergies between bees, wasps, ants (SP6) and decomposers across the tree diversity gradient (SP1). WP3 tests the hypothesis that nests closer to the ground and nests allowing access for ants have low bee and wasp occupancy with little parasitism rates. This is because bee nests at suboptimal nesting conditions are described to be enemy-released. How the presence of decomposers influence the presence of bees, wasps and their parasitoids (e.g. through ecological engineering of wood-boring beetles), with and without ants, along the tree diversity gradient will be part of SP1.
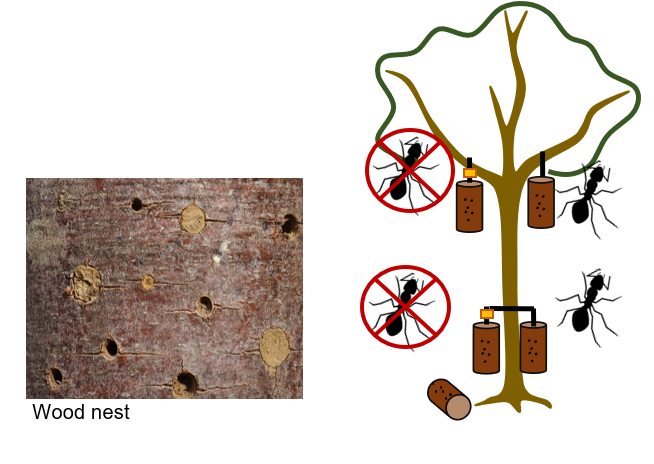
Researchers
Principle Investigators:
Alexandra-M. Klein, University of Freiburg
Felix Fornoff, University of Freiburg
Manuela Sann, University of Hohenheim
Doctoral Researcher:
Massimo Martini, University of Freiburg
Chinese Partner:
Arong Luo, Institute of Zoology, Chinese Academy of Sciences, Beijing
Partner:
Michael Orr, Naturkundemuseum Stuttgart
SP5: Trophic interactions across tree regeneration stages
Objectives
SP5 addresses key questions at the bridge of functional biodiversity, population and community ecology research in forests by combining information from observational and experimental approaches. Objectives are related to mechanisms of woody recruitment of early reproducing tree species in BEF-China. Referring to findings from the BEF-China project that indicated strong multitrophic regulatory influence on tree seedling establishment already in the early stage of the experiment, we intend to widen the perspectives to two directions. While, first, we aim to increase the number of multitrophic interactions studied against the background of JC effects across multiple stages of the tree regeneration life cycle, we, secondly, do so along the gradient of tree diversity. Our main objectives are to quantify the contribution of tree diversity on (i) the reproductive cycle stages of trees that are particularly affected by invertebrate interactions, (ii) pre- and post-dispersal seed predation and (iii) germination and seedling herbivory as trophic interactions that affect these reproductive cycle stages as well as (iv) the contribution of generalist and specialists to these trophic interactions.
BEF-China provides the unique possibility to assess these relationships by making use of the full gradient of tree diversity established along the core plots and VIPs.
We will address the major objectives organised in three work packages:
1) Demography of early life stages:
to assess natural tree regeneration (1) for a selected set of tree species in the experiment along the full tree diversity gradient on the observational level,
2) Key process of seed predation:
a) to quantify pre-dispersal seed predation (2) along the full gradient of tree diversity established by experimental means
b) to quantify dispersal and post-dispersal seed predation by means of invertebrate exclosures (3)
3) Key process of herbivory:
to assess germination and seedling herbivory experimentally by quantifying JC effects along the tree diversity experiment (4, 5)
Researchers
Principle Investigators:
Alexandra Erfmeier, University of Kiel
Tim Diekötter, University of Kiel
Doctoral Researcher:
Noga Abecassis, University of Kiel
Chinese Partner:
Zhi-Shu Xiao, Institute of Zoology, Chinese Academy of Sciences, Beijing
SP6: Ant trophic interactions and functions
Objectives
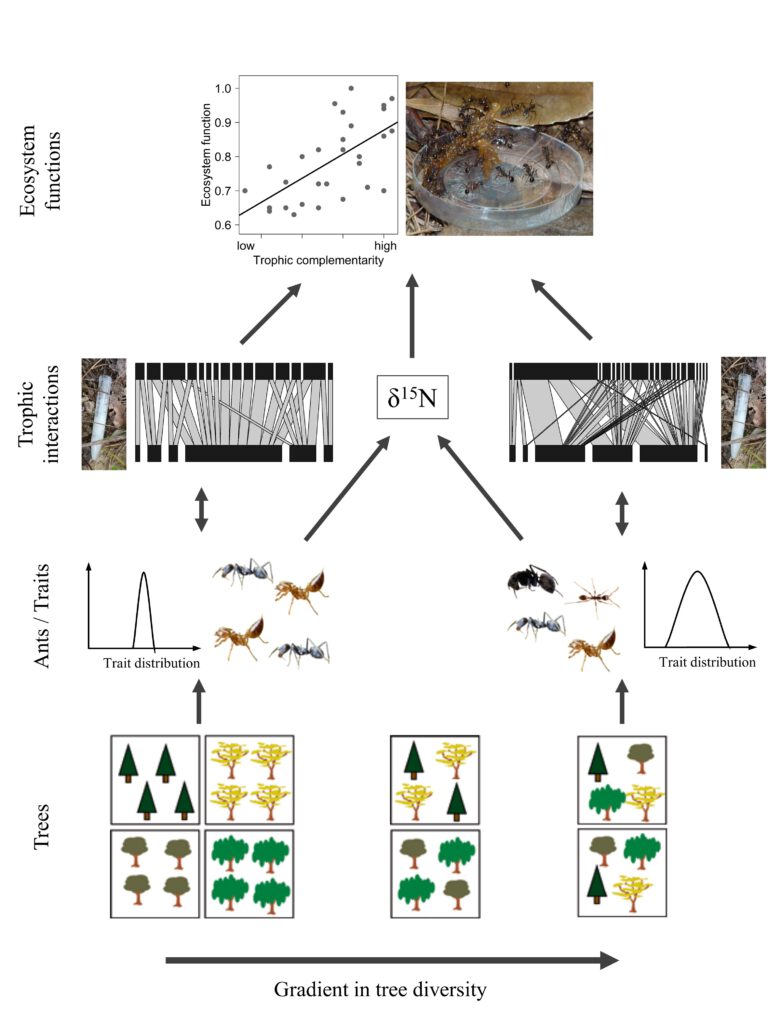
The overall objectives of SP6 are to test if experimentally manipulated tree diversity influences the diversity, trait distribution, trophic interactions and ecosystem functions of ants (Fig. 1). We identify three interdependent main aims that will be the leading themes of the work packages (WP) and jointly allow to address the objectives: (1) Quantify the diversity and functional trait distribution of ants in all 300 core plots; this will not only allow to functionally characterise all collected ant species and to test if ant diversity and traits change with tree diversity, but also provide important baseline data for our other objectives and deliver a generic diversity variable available to other SPs and synthesis. This objective also incorporates ant specimens collected by SP1 and SP3. (2) Identify the trophic position and assess trophic interactions of ants by performing resource-choice experiments in the 300 core plots and by measuring δ15N from specimens in the 64 VIP plots; with these data we can calculate trophic redundancy, trophic niche breadth on species and community level, and infer whether the trophic interactions of single species and the entire ant community are related to tree diversity. We will also test which traits relate to trophic niche breadth and relative trophic positions. Data on preferences for acronutrients (from resource-choice experiments) will feed-back to the stoichiometry projects (SP2, SP3) and allow for a more complete quantification of nutrient limitations and element flows. (3) Measure the ecosystem function “scavenging” in the VIP plots and test how the interplay between tree diversity, ant diversity, trait distribution and trophic interactions influences ecosystem functionality. This third objective will build upon the other two objectives and integrate across components of biological organization to link diversity and interactions to a function.
Researchers
Principle Investigators:
Michael Staab, University of Darmstadt
Heike Feldhaar, University of Bayreuth
Doctoral Researcher:
Joshua Spitz, University of Darmstadt
Chinese Partner:
Xiaojuan Liu, Institute of Botany, Chinese Academy of Sciences, Peking
Z2: : Data management and synthesis
Objectives
The overall objectives of Z2 are to
- ensure efficient data management and promote synergies in the processing of data among MultiTroph’s SPs through extensive services provided by training courses and the maintenance and ongoing development of the BEF-China data portal, as well as supporting data and code open access publication strategy,
- develop early synthesis connecting MultiTroph’s SPs based on available and newly generated data that link up network structure, multidiversity, and multifunctionality. Multidiversity assembly and trophic interactions need time to establish. For this reason, including previous data from BEF-China is essential.
The main aims and guiding hypotheses are:
- Continue and update data portal
In preparation for MultiTroph, we have updated and migrated the BEFdata portal from Leipzig to Halle University (https://data.botanik.uni-halle.de/bef-china/). Halle University has provided the required server capacity and technical surrounding to run the portal. Among other features, the BEFportal now has a flexible design and can work with screens of all sizes (e.g. tablet, mobile phone). In addition, we have implemented an improved data search. On Github, about 30,000 lines of code have been added and large parts of the old code have been rewritten (https://github.com/befdata/befdata). While the data of the BEF-China projects (the former Research Unit FOR 891 “BEF-China”, the TreeDì graduate school and numerous Chinese projects) are curated together, their access and re-use is organized in different subprojects, allowing separate access rules and proposals.
The main aim (1) of MultiTroph’s data management is to provide data management along the entire data lifecycle. However, as the structure of the database has been updated, we will focus on improving functionality, such as developing tools for extracting and merging data for synthesis datasets, thus leveraging reproducible scientific analyses. We will work closely with the National Research Data Infrastructure (NFDI) initiative, particularly with NFDI4Biodiversity (https://www.nfdi4biodiversity.org/) focusing on biodiversity data, to which we are connected through iDiv. (2) We will provide reproducible workflows into which the particular data analyses will be integrated. (3) We will offer regular training activities on using the data portal to support all activities of the data life cycle (e.g. data generation, documentation, building pipelines, archiving and publishing). Finally (4), we will work to combine paper proposals, which are a key element in BEFdata, with applications that are required for working on the BEF-China platform and which are administered by the BEFChina steering committee. Paper proposals offer a safe environment to share data to propagate data ownership through scientific workflows. - Analyse multitrophic network structure and multidiversity
We anticipate that tree diversity effects on multidiversity of higher trophic levels are to a large part indirect and mediated by the structure of interaction networks linking producer and higher trophic levels. We hypothesize that increasing tree species richness increases trophic complementarity and affects further network characteristics through specialization of network partners. We expect increasing specialisation with increasing producer richness as a general mechanism of trophic networks, which in turn promotes higher trophic level multidiversity. However, differences may be expected for antagonistic versus mutualistic networks, and more mobile groups (with their ability to utilize resources across larger spatial scales) and higher trophic levels (often less affected by changes in plant diversity) are hypothesized to show less pronounced relationships with the manipulated tree communities. - Link network characteristics and multidiversity to ecosystem (multi)functionality
In general, we expect individual ecosystem functions and ecosystem multifunctionality to be promoted by higher diversity across trophic levels. We hypothesize that cascading effects of tree diversity via interaction network structure and multidiversity will also play important roles in modifying process rates and the stability of ecosystem functions. The degree of generalization and specialization in the interaction webs will strongly determine the outcome of such cascading effects: specialization of network partners increases the importance of trophic complementarity in explaining ecosystem functioning. Trophic complementarity may increase with increasing multitrophic diversity. In contrast, a strong dominance of generalist (highly connected) species might lead to less pronounced effects of diversity, as generalists might maintain high functionality by occupying a broader niche space already at lower diversity. Finally, we expect network structure to also influence the variability of ecosystem functions, with particularly functional redundancy within the networks (e.g. high average number of consumers per host, i.e. community-level vulnerability) reducing variability in functions across space and/or time.
Researchers
Principle Investigators:
Andreas Schuldt, University of Göttingen
Helge Bruelheide, University of Halle
Postdoctoral Researcher:
Georg Albert, University of Göttingen
Chinese Partner:
Huijie Qiao, Institute of Zoology, Chinese Academy of Sciences, Beijing
Xiaojuan Liu, Institute of Botany, Chinese Academy of Sciences, Beijing
Yi Li, Institute of Botany, Chinese Academy of Sciences, Beijing